StackWave is thrilled to announce the release of Affinity v16, which brings support for Next Generation Sequencing (NGS) data analysis. We have prioritized NGS support in v16 in response to feedback from discovery scientists that need better support for identifying novel antibodies with therapeutic potential from their B cell sequence data. Affinity v16 includes an intuitive and robust interface to import and analyze NGS sequence data, identify clones of interest based on enrichment, mutation frequency, and amino acid properties, support alignment of clonal sequences, export the set of sequences for external use, and request expression for selected clones of interest.
Automatically perform NGS analysis on sequence databases
Load your NGS data files, tell Affinity a little bit about them, and the system will automatically analyze your sequences and prompt you to review the results.
Identify Clones of Interest
Review the analysis of your sequences, identify clones of interest by reviewing the automatically-generated enrichment, mutation frequency, and amino acid properties, or perform sequence alignment on clones and then group those clones into sets.
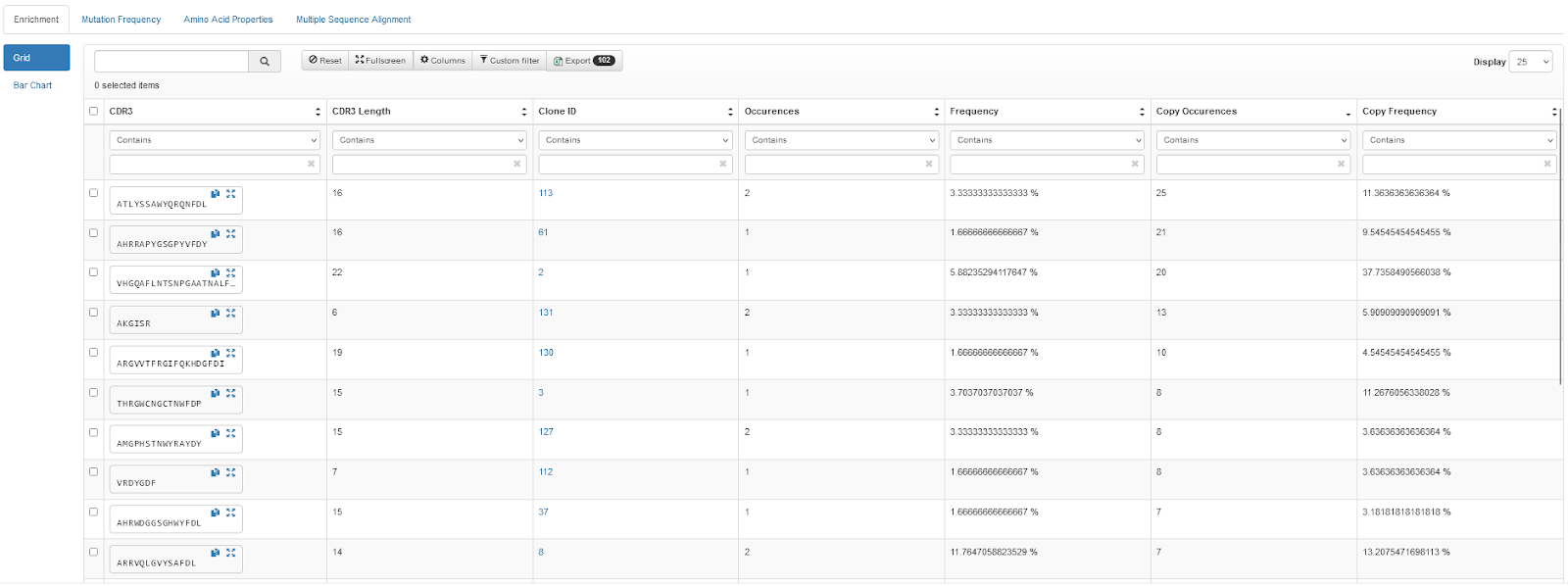
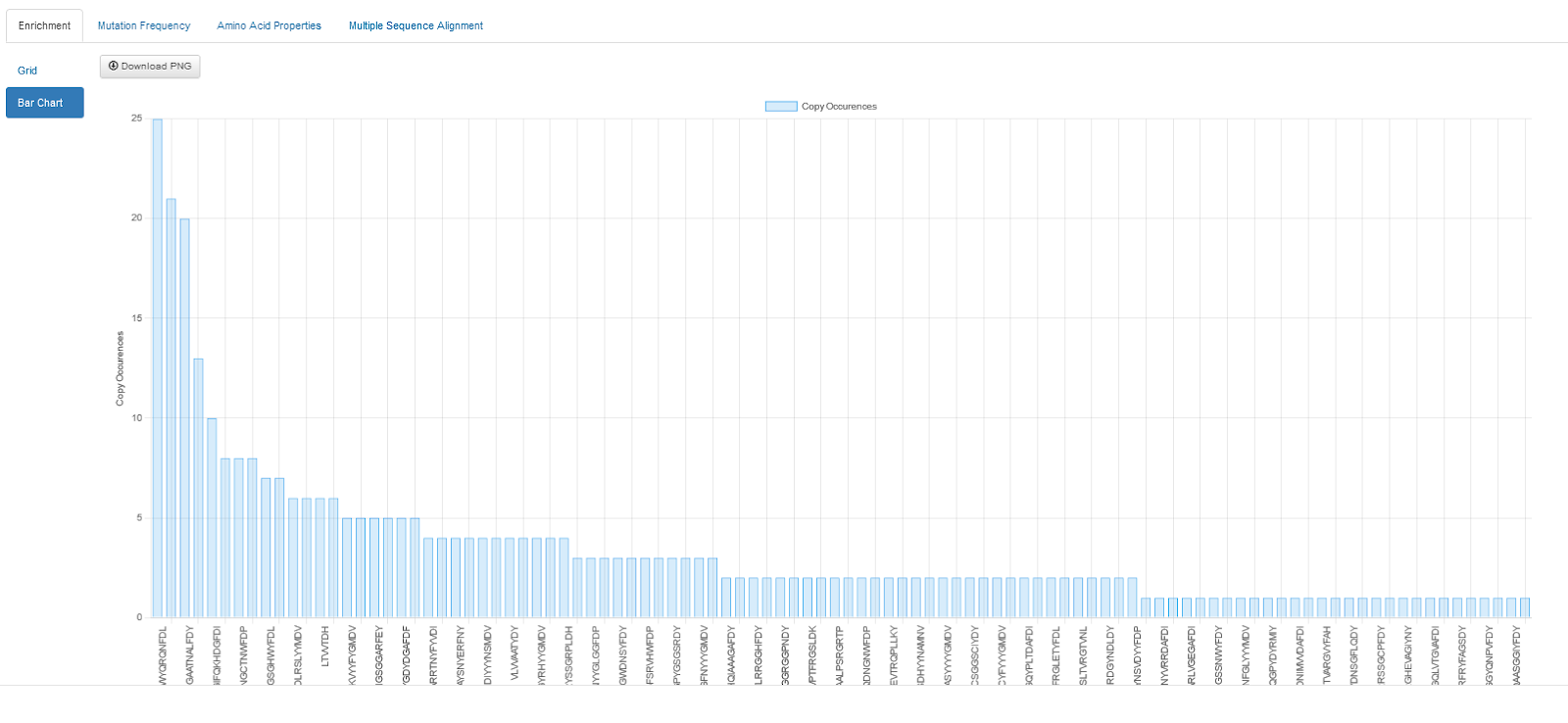
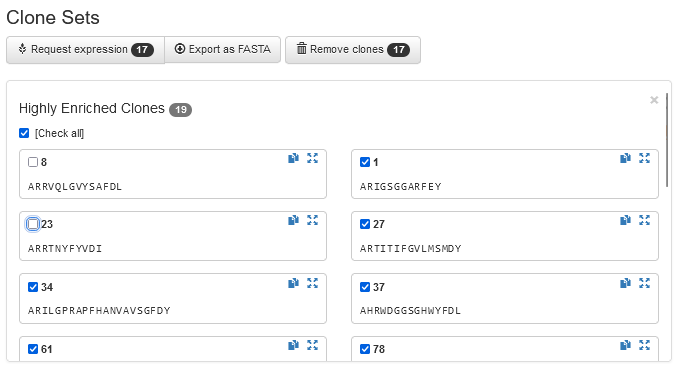
Request Production and Export Sequences
A set of your most interesting clones can be exported for external use at the click of a button, or sent to the built-in Protein Production workflow, which will facilitate and track every step of the vector construction and protein expression process.
We are incredibly excited to bring these new tools to our users. Please don’t hesitate to reach out to us at inquiries@stackwave.com for a free trial or demo of StackWave Affinity!