Sequence alignment allows users to enter a series of sequences and identify commonalities or differences between them
Sequences can be loaded either by loading a FASTA file or by manually entering the sequences in the UI.
Click Submit to process the sequences for alignment.
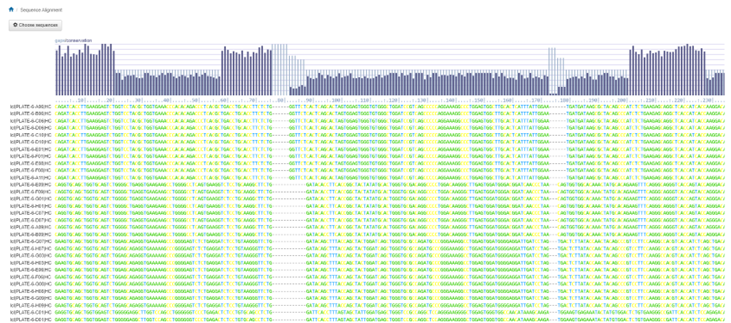
The resulting interface offers a visual representation of the gaps and conservation found during analysis of the sequences. Each sequence is displayed below the gaps/conservation display, ordered by the sequences which had the highest alignment at the top.
