Users can register proteins in one of two ways, either from sequences or from plasmids.
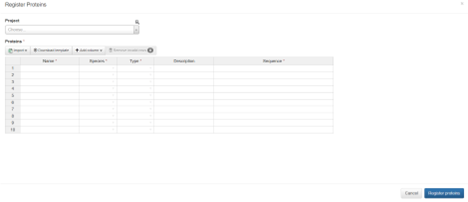
Select the desired registration option to open the registration form. Users can optionally select a project, and can either enter their data manually in the form or import an XLS file to quickly register large numbers of proteins.
The default fields on protein registration are Name, Species, Type, Description, and Sequence (or Plasmid, depending on registration type), however the user is able to add any number of additional sequences/plasmids if desired.
Click Register Proteins in the bottom right to complete the process and be redirected back to the Proteins index page.
Protein Index and Details
The index page for proteins allows users to review the existing proteins in the system. Filters can be applied on any column to narrow the shown proteins. Click on any protein ID or name to navigate to its details page.
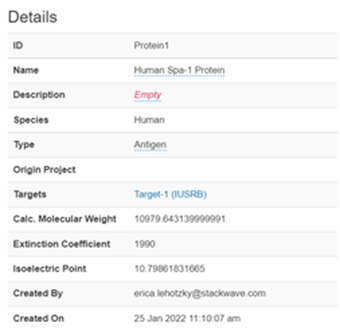
This page displays all the details of the protein, its associated lots, assay results, and sequence information.
The details section will allow users with appropriate permissions to edit the name, description, and type of the protein, and displays the calculated values molecular weight, extinction coefficient, and isoelectric point.
In the upper right of the page, the user is able to tag, comment, and attach any files to the entity as needed.
The Lots section allows the user to review any lots which have been registered of the protein being viewed, as well as register new proteins. To do so, click the Register Lots button. This form allows you to manually enter a volume, concentration, vendor, vendor lot ID, buffer, and modification. To navigate to a lot’s detail page, click on the Lot ID.
Beneath Lots are the Assay Results - this section allows the user to select any assay type, and then any results which have been registered against lots of this protein of the selected type will be displayed.
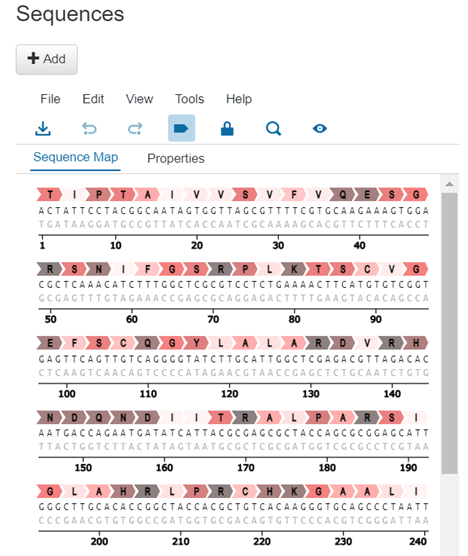
Finally, the details page will display the Sequences associated with the plasmid. The DNA and amino acid sequence will be displayed, and the tool can be used for a variety of sequence related activities, such as exporting, translation, and more. Users can also add new sequences in this section by clicking Add and entering an amino acid sequence.