Registration of clone plates is performed on the Clone Plates page of the Discovery module
Often, after creation of an initial set of clone plates, a series of assays will be performed whose results will rule out certain samples. The clones which remain will be rearrayed into a new set of plates. Affinity facilitates this process through the Rearray features, also found on the Clone Plates page.
Clone Plates
To register a set of plates, click on Register Clone Plates in the Clone Plates page, and then select either Selection or Hybridoma. Enter the project, target, number of plates, transfer pattern, and samples - in the case of Hybridoma, the available samples will be tissue samples registered in Affinity, whereas if the clone plates are based on Selection, the user will be asked to specify the Type (Bacteria or Phage), and the samples available for selection will be those samples which resulted from the Selection process.
In both cases, the system will automatically register the number of plates specified - each plate is assigned a Plate ID, its designation will be Registration (as opposed to derived from Rearray), and the Source will indicate whether the plates came from Selection or Hybridoma.
Clicking on the Plate ID will direct the user to the Inventory Browser, which shows the plates location in inventory (the default location can be altered by administrators), as well as details of the samples in each well and the ability to export a plate map.

Returning to the Clone Plates page, the Duplicate button creates an exact copy in inventory of each plate selected - the only difference will be the Plate ID and the Created On timestamp.
Rearray Plates
Often users will need to progressively narrow the contents of their clone plates based on the results of various assays or according to sequence alignment. To facilitate this process in Affinity, use the Rearray Selected Plates button on the Clone Plates page.
Select one or more plates using the checkboxes in the Clone Plates index grid and then click Rearray Selected Plates. This will open the Rearray Clones modal.

Select a phase, source pattern, and transfer pattern, which will direct the layout of the resulting plates. In Sample Replication, the user can specify whether to fill each plate with a source sample - for example, if there are only 3 samples included after applying filters, the result of the rearray will be three monoclonal plates where every well (excluding controls) contains a single sample. Enter a value in Plate Copies to create one or more copies of each plate resulting from the rearray. There are two means of filtering clones in the source plates, the first being Assay Data and the second being Alignment.
Assay Data
To filter clone plates by assay data, the user should first load a set of assay results. This can be done using the “Submit Assay Results” button on the Rearray Clones page, the “Submit Assay Results” button in the navigation bar at the top of any page in Affinity, or on the Assay Results page of the Analytics module.

Clicking “Add Result Set” will open a modal which allows the user to select which type of data to load for the clone plates previously selected and specify which columns to load into the grid.
Users can load multiple result sets at once, including loading the same type of data if necessary.
Once the assay results are loaded, use the filtering tools to apply desired filters. See example below.

Users can also use the standard Affinity grid Custom Filter to create as complex filter as needed - see example below.

The user can use the Save Criteria/Load Criteria buttons in order to quickly retain and recall complicated filters which will be reused.
Alignment
Filtering by Alignment allows the user to select a particular sequence region and then view how the sequences of the clones under study line up. The user can then select specific clones to manually exclude from the rearray results. This tool is often useful when the user wants to reduce overly similar sequenced clones in their clone plates.

Rearrays
Clicking Rearray Samples will create a new sample set which includes a number of plates necessary for the resulting clones. On the Sample Sets page, you can view various data surrounding the source of the sample set, including whether the source was from selection or hybridoma, the specific plates used to create the set, phase, creator, and date created. In addition, any filters set on the Rearray Clones page or specifically excluded samples if filtering by alignment and plates which resulted will be displayed as well as the samples contained within those plates.
The rearrays index page in the Discovery module contains all sample sets created through rearray if the user needs to revisit the method which resulted in any rearrayed clone plates.

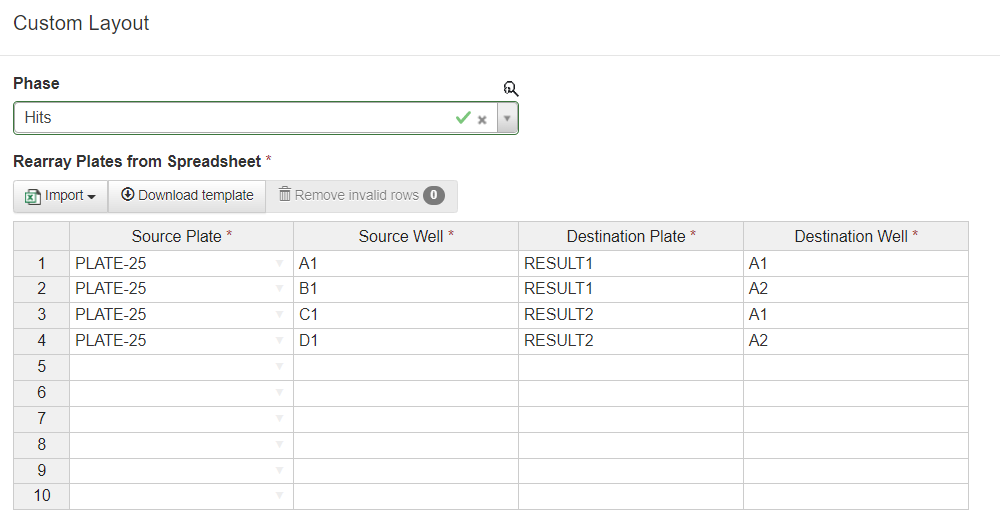
Rearray (Custom Pattern)
Occasionally, a rearray requires a very specific mapping from source to destination plates which cannot be systematically described in the standard Rearray Selected Plates interface. In this case, the Rearray (custom pattern) tool should be used. This allows the user to explicitly describe where each source plate and well should be directed. The value entered in the Destination Plate field will be applied as the name of the resulting plate (although a standard Plate ID will still be generated) - entering different values in the Destination Plate field will result in that number of plates being generated. For example, in the below image, the rearray will result in two plates named RESULT1 and RESULT2, each with a sample in wells A1 and A2.
Generate Supernatant Plates
When performing assays against supernatant plates, use the "Generate supernatant plates" tool. This tool will duplicate the selected plates, however the resulting plates have a unique feature - assay results loaded against their samples are applied as well to the corresponding samples in the originating plate from which they were generated. This allows a user to generate the labels for their assay plates and still view the results of those assays against the original plates.
