Let's walk through an example nanobody campaign with StackWave Affinity™
Establishing a goal
 Our goal is the identification of a handful of nanobodies with a range of affinities against multiple epitopes of two commonly co-expressed tumor-associated antigens (TAA). This will allow us to experiment with different trimeric formats incorporating an anti-CD3 fragment to try and enhance tumor cell killing by cytotoxic T cells. We've built several phage display libraries for the purpose: one sourced from camelids, one from sharks, and a synthetic library that tests a huge array of CDR loop lengths. We'll cover registration and use of these libraries shortly. We've also purchased tubes of our TAA targets, which we can register as Protein materials that each have a Lot containing multiple tube-based Samples. With our reagents registered, we'll register our two targets in Affinity against their Protein records and receive auto-generated IDs of Target-1 and Target-2.
Our goal is the identification of a handful of nanobodies with a range of affinities against multiple epitopes of two commonly co-expressed tumor-associated antigens (TAA). This will allow us to experiment with different trimeric formats incorporating an anti-CD3 fragment to try and enhance tumor cell killing by cytotoxic T cells. We've built several phage display libraries for the purpose: one sourced from camelids, one from sharks, and a synthetic library that tests a huge array of CDR loop lengths. We'll cover registration and use of these libraries shortly. We've also purchased tubes of our TAA targets, which we can register as Protein materials that each have a Lot containing multiple tube-based Samples. With our reagents registered, we'll register our two targets in Affinity against their Protein records and receive auto-generated IDs of Target-1 and Target-2.
Automating reports against the goal
To simplify the organization of our nanobody campaign's data, we'll create a new project in Affinity and set up some automated reporting. We'll call this new project "Nanobody Discovery Campaign." Creating the project takes us to a new page that collects all of the information about the project - to date just the name and auto-generated project ID. However, we can click the "Create Dashboard" button to create a new reporting dashboard that will automatically associate with and display on this page. We'll call this our "Nanobody Campaign Dashboard."
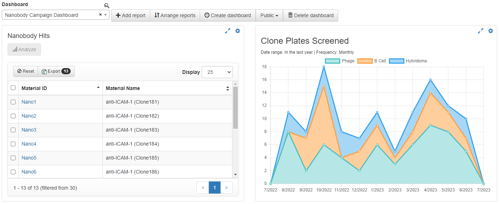
For now, we're simply going to add a Therapeutic Goals Monitor to our dashboard. The Project field is automatically populated on our behalf, but for our Therapeutic Goals Monitor we'll need to apply filters for the type of therapeutic (Nanobody), the criteria for our therapeutic (range of Kd values from our Kinetics assay corresponding to at least three different Bins from our Binning assay, all with unique heavy chain CDR3 sequence elements), and a set of email addresses to be notified when the goal is reached. For the Status Report, we again filter to Nanobodies, but we also set the report columns to Kinetics, Binning, ELISA, and heavy chain CDR3 sequence data. After creating the dashboard, the project page will update to include the new dashboard; the monitor will be shaded red since the goals have not been met, and the Status Report will be empty.
Executing the nanobody discovery process
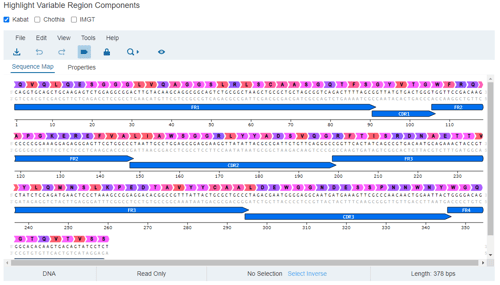
Before we begin our campaign, to realize the full value of Affinity's sequence analysis software, we first ensure the germline sequences associated with our nanobody libraries are registered with the system by either uploading FASTA files for the V(D)J genes or by registering germline sequences using Affinity's in-browser spreadsheet. We then register our libraries and associate the appropriate germline sequences with them. When we eventually upload our clone sequences, Affinity will compare them against the supplied germline sequences to identify their origin and CDRs.
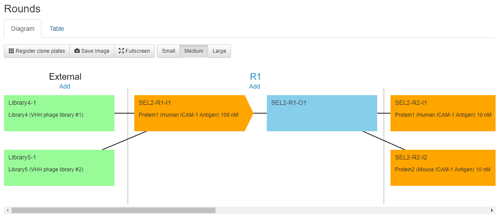
We're now ready to begin phage panning. We'll create two Selection efforts in Affinity, one each for Target-1 and Target-2, and each of which is associated with our three Libraries as inputs and the appropriate Protein that we plan on using. Each Selection effort then loads with our three libraries available under the "External" column of the panning diagram. From here, we can create multiple per-round inputs and outputs that describe our panning protocol and connect the panning effort, round, or individual selection arms with corresponding notes in our ELN.
At the end of our panning effort, we use NGS to sequence each of our final pools of phage. The assembled reads are then fed into Affinity's NGS module, where they are analyzed using the previously-provided germline sequences, scanned for potential development liabilities, and grouped into clonotypes. Affinity's NGS visualization tools then allow us to explore this data to find the most highly-enriched, liability-free clones. From each phage panning effort we want to fill three 96-well plates for high-throughput expression and initial characterization - these will be labeled as our Hits and become the initial set of Nanobodies that we screen for therapeutic lead potential. We select the clones of interest and click the "Request expression" button to simultaneously register them as Hits and submit them to our protein expression team for high-throughput expression.
Affinity supports every step of the cloning and expression processes, allowing molecular biology and protein expression teams to manage all of the work required to produce our plates of nanobodies. You can read more about the entire process here, but for the purpose of this guide we'll skip ahead to the part where we receive the plates of expressed protein and begin to assay their contents.
Examining the characterization data
Affinity can read from a large (and growing) library of instrument formats, including those produced by the Carterra LSA. We can submit our Carterra kinetics and binning results files directly to Affinity for processing via the "Submit Assay Results" button available on every page. Once these files have been processed, the dashboard on the Nanobody Discovery Campaign's page will update to reflect the new data. In this case, our goals monitor has populated with nanobodies that meet our criteria, indicating that we have the number of hits we were looking for. We can review the specifics of the data in our Status Report and use this report to select the nanobodies we want to promote to leads. Clicking the "Promote" button at the top of the Status Report marks the selected nanobodies as our leads for this campaign.
With our leads in hand, we can begin to generate our trimers for a new round of characterization and functional testing.
StackWave Affinity™ Advantage
Single Data Repository
One source of truth for program data allows you to review results and look for patterns within teams, projects, and across the organization.
Workflow Automation
Reduce repetitive tasks and human error by automating common antibody discovery workflows, data transformations, and analyses.
Support for Every Team
Simplify collaboration and communication between antibody discovery, molecular biology, protein expression, and analytics teams.
Custom Dashboards
Monitor progress in real time to identify bottlenecks as they arise and derive data-driven, actionable insights with advanced reporting.
“StackWave gave us confidence in our leads by collecting all of the data by our potential therapeutics in one place and making that data actionable by allowing us to compare antibodies of interest."
"I worked with StackWave for ~4 years at my previous job at Magenta Therapeutics to implement our LIMS. We put in place an incredible system for our entire workflow, from plasmid registration to in-vivo study data registration. Andrew and his team are very experienced and knowledgeable and helped us deliver a user-friendly interface for our scientists."

